Research Highlights
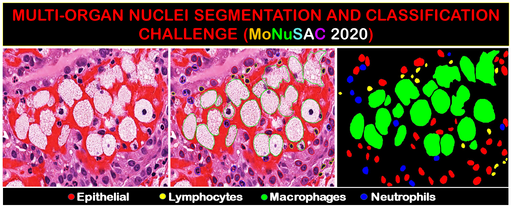
We created the large and diverse dataset of nucleus boundary annotations and class labels to advance state-of-the art in generalized multi-class nuclei segmentation. We organized an international competition to train deep neural networks to segment and classify nuclei in digital pathology at ISBI 2020. Detecting various types of cells in and around the tumor matrix holds a special significance in characterizing the tumor micro-environment for cancer prognostication and research. This work is under review in IEEE Transactions on Medical Imaging. [Workshop]
Generalized Nuclei Segmentation
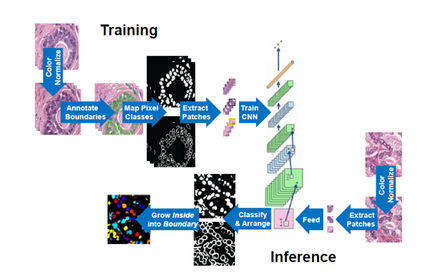
We developed a deep learning based generalized nuclear segmentation algorithm and also released a hand-annotated benchmark nuclear segmentation dataset encompassing multiple organs, patients and disease states. Additionally, we proposed a metric to evaluate nuclear segmentation results that penalizes both object- and pixel-level errors in a unified manner. To facilitate state-of-the art in generalized nuclei segmentation, we co-organized an international competition to segment nuclei in digital pathology in MICCAI 2018. Nuclei segmentation algorithms aid in several downstream computational pathology tasks including cancer mutation prediction, cancer staging, treatment effectiveness and outcome prediction. This work recently got published in IEEE TMI [Paper].
Co-existence of Multiple Subtypes of Breast Cancer in a Single Patient
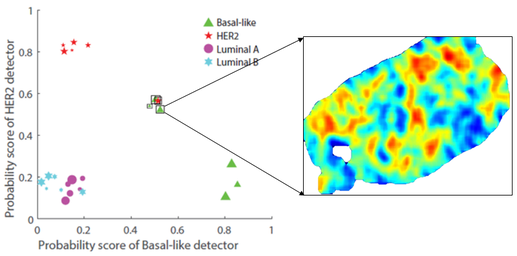
Breast cancers are subtyped and treated differently based on their molecular makeup. Even within a given subtype, some cancers are admixed with other subtypes. We developed a method to detect
co-existence of two common sub-types of breast cancer –HER2 and basal-like – in whole slide images of H&E-stained tumors in the same patient. [Paper]
co-existence of two common sub-types of breast cancer –HER2 and basal-like – in whole slide images of H&E-stained tumors in the same patient. [Paper]
Morphologic Basis of the Radiomic Features, prognostic of progression-free survival, in Glioblastoma
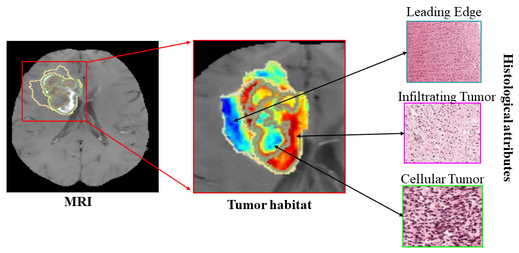
Developed a method to create a radiomics risc score to stratify patients into low and high-risk groups of poor progression-free survival in response to standard-of-care treatment in Glioblastoma. Radiomics risc score was created using quantitative features obtained from different sub-compartments of the tumor habitat. We then identified significant morphologic associations of the prognostic radiomic features on pretreatment MR images (used to create the RRS) with corresponding pathologic attributes from H&E stained digitized tissue images to assess the tumor phenotypical traits responsible for tumor invasion and treatment resistance. [Paper]
Prostate Cancer Recurrence Prediction
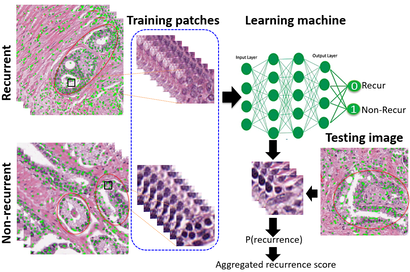
Accurate prediction of the treatment outcome is important for cancer treatment planning. We developed a fully-automated deep learning algorithm to predict prostate cancer recurrence after radical prostatectomy from H&E stained tissue images of individual patients. We demonstrated that our algorithm outperforms the current clinical grade diagnostic protocol to assess mid-Gleason grade prostate cancer. Our approach might help in choosing between a combination of treatment options such as active surveillance, radical prostatectomy, radiation, and hormone therapy. [Paper]
Publications
Journals
Abstracts
- Neeraj Kumar, Daniel Skubleny, Michael Parkes, Ruchika Verma, Sacha Davis, Luke Kumar, Amira Aissiou, and Russell Greiner, ``Learning Personalized Survival Models from PanCancer Whole Transcriptome Data", in Clinical Cancer Research, 2023.
- Shi-ang Qi, Neeraj Kumar, Ruchika Verma, Jian-Yi Xu, Grace Shen-Tu, and Russell Greiner, ``Using Bayesian Neural Networks to Select Features and Compute Credible Intervals for Personalized Survival Prediction", in IEEE Transactions on Biomedical Engineering, 2023.
- Sarthak Pati et. al., ``Federated Learning Enables Big Data for Rare Cancer Boundary Detection", in Nature Communications 13, no. 1 (2022): 1-17.
- Sukanya Iyer et. al., ``Novel MRI deformation-heterogeneity radiomic features are associated with molecular subgroups and overall survival in pediatric medulloblastoma: Preliminary findings from a multi-institutional study." Frontiers in Oncology 12 (2022): 915143.
- Ruchika Verma et. al., ``Stable-and-discriminatory radiomic features from the tumor and its habitat associated with progression-free survival in Glioblastoma: A multi-institutional study", in American Journal of Neuroradiology, 43.8 (2022): 1115-1123.
- Neeraj Kumar, Ruchika Verma et. al., ``Computer Extracted features of Nuclear Morphology on H\&E images distinguish Stage II and IV Colon Tumors", in The Journal of Pathology (2022), 257.1 (2022): 17-28.
- Ruchika Verma et. al., "MoNuSAC2020: A Multi-organ Nuclei Segmentation and Classification Challenge", in IEEE Transactions on Medical Imaging (2021).
- Ruchika Verma, et. al., “Tumor-habitat derived radiomic features that are prognostic of progression-free survival in Glioblastoma are associated with key morphologic attributes on pathology: A feasibility study”, in Radiology: Artificial Intelligence, Vol. 6, no. 2, Nov 2020.
- Neeraj Kumar, Ruchika Verma, et al., "A Multi-organ Nucleus Segmentation Challenge", in IEEE Transactions on Medical Imaging, Vol. 39(5), Pages 1380-1391, Oct 2020.
- Ruchika Verma*, Sarthak Pati* et. al., "Reproducibility analysis of multi-institutional paired expert annotations and radiomic features of the Ivy Glioblastoma Atlas Project (Ivy GAP) dataset" in Medical Physics Oct 2020 (* indicates equally contributing first author).
- Niha Beig, Kaustav Bera, Prateek Prasanna, Jacob Antunes, Ramon Correa, Salendra Singh, Anas Saeed Bamashmos, Marwa Ismail, Nathaniel Braman, Ruchika Verma, et. al., "Radiogenomic-based survival risk stratification of tumor habitat on Gd-T1w MRI is associated with biological processes in Glioblastoma: A multi-institutional study", in Clinical Cancer Research, Feb 2020.
- Amir Reza Sadri, Andrew Janowczyk, Ren Zhou, Ruchika Verma. et. al., "MRQy: An Open-Source Tool for Quality Control of MR Imaging Data", in Medical Physics, Nov 2020.
- Neeraj Kumar, Ruchika Verma, Sanuj Sharma, Surabhi Bhargava, Abhishek Vahadane and Amit Sethi, "A dataset and a technique for generalized nuclear segmentation in histological images for computational pathology", in IEEE Transactions on Medical Imaging, vol. 36, no. 7, pp 1550-1560, July 2017.
- Neeraj Kumar, Ruchika Verma and Amit Sethi, "Convolutional neural networks for wavelet domain single image super resolution", in Pattern Recognition Letters, Volume 90 Issue C, Pages 65-71, April 2017.
- Ruchika Verma et. al., ``Sex-specific Computational Histopathological Signatures Prognostic of Overall Survival in High-Grade Gliomas via Deep-learning: Large Multi-cohort, Retrospective Study”, in Science Advances.
- Olivia Krebs, German Corredor, Jeffrey Helgager, Prerana Bangalore Parthasarathy, Tyler Alban, Ruchika Verma , Justin Lathia, Manmeet Ahluwalia, and Pallavi Tiwari, ``Sex-specific pathological markers related to the spatial organization of tumor infiltrating lymphocytes predict overall survival in high-grade gliomas”, in Journal of Neuro-Oncology.
- Pallavi Tiwari and Ruchika Verma, "The Pursuit of Generalizability to Enable Clinical Translation of Radiomics", in Radiology: Artificial Intelligence.
- Alvaro S. Garzon, Ruchika Verma et. al., "A Hierarchical Deep Learning Approach for Segmentation of Glioblastoma Tumor Niches on Digital Histopathology", in Medical Imaging 2022: Digital and Computational Pathology, Vol. 12039. SPIE, 2022.
- Yashas Hiremath, Marwa Ismail, Ruchika Verma, Ashish Gupta, and Pallavi Tiwari, "Combining Deep and Hand-crafted MRI features for identifying gender-specific differences in ASD versus normal controls", in SPIE Medical Imaging 2020: Computer-Aided Diagnosis. Vol. 11314. SPIE, 2020.
- Marwa Ismail, Ramon Correa, Kaustav Bera, Ruchika Verma, et. al , "Spatial-And-Context aware (SpACe) "virtual biopsy" radiogenomic maps to target tumor mutational status on structural MRI" in Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, October 4–8, 2020, Proceedings, Part II 23. Springer International Publishing, 2020.
- Ruchika Verma, Ramon Correa, Virginia Hill, Niha Beig, Abdelkader Mahammedi, Anant Madabhushi and Pallavi Tiwari, "Radiomics of the lesion habitat on pre-treatment MRI predicts response to chemo-radiation therapy in Glioblastoma", in Medical Imaging 2019: Computer-Aided Diagnosis. Vol. 10950. SPIE, 2019.
- Niha Beig, Prateek Prasanna, Virginia Hill, Ruchika Verma, Vinay Vardan, Anant Madabhushi and Pallavi Tiwari, "Radiogenomic characterization of response to chemo-radiation therapy in Glioblastoma is associated with PI3K/AKT/mTOR and apoptosis signaling pathways", in Medical Imaging 2019: Computer-Aided Diagnosis. Vol. 10950. SPIE, 2019.
- Sukanaya Iyer, Marwa Ismail, Benita Tamrazi, Ashley Margol, Ruchika Verma, et al., "Deformation heterogeneity radiomics to predict molecular subtypes of pediatric Medulloblastoma on routine MRI", in Medical Imaging 2019: Computer-Aided Diagnosis. Vol. 10950. SPIE, 2019.
- Neeraj Kumar, Ruchika Verma, Ashish Arora, Abhay Kumar, Sanchit Gupta and Amit Sethi, "Convolutional neural networks for Prostate cancer recurrence prediction", in Medical Imaging 2017: Digital Pathology. Vol. 10140. SPIE, 2017.
- Ruchika Verma, Neeraj Kumar, Amit Sethi and Peter H Gann, "Detecting multiple sub-types of breast cancer in a single patient", in IEEE International Conference on Image Processing (ICIP). IEEE, 2016.
Abstracts
- Ruchika Verma, Wei Wu, Neeraj Kumar, Elizabeth Yu, Won-Tak Choi, Sarah Umetsu, and Trever Bivona, "Deep learning-based integration of esophageal cancer morphology with genomics" AACR Annual Meeting 2021.
- Ruchika Verma, Mark Cohen, Paula Toro, Mojgan Mokhtari, Pallavi Tiwari, "Predicting Overall Survival in Glioblastoma using Histopathology via an end-to-end Deep Learning pipeline: A large multi-cohort study", Annual Meeting Society for Neuro-Oncology, 2021.
- Ruchika Verma, Yasmeen Rauf, Ipsa Yadav, Volodymyr Statsevych, Jonathan Chen, Wendi Evanoff, Manmeet Ahluwalia, Pallavi Tiwari, "Radiomic features from lesion habitat predict response to combination of Nivolumab and Bevacizumab in patients with recurrent Glioblastoma: A feasibility study", Annual Meeting Society for Neuro-Oncology, 2021.
- Ruchika Verma, Yasmeen Rauf, Ipsa Yadav, Volodymyr Statsevych, Jonathan Chen, Wendi Evanoff, Manmeet Ahluwalia, Pallavi Tiwari, "Prediction of response to combination of Nivolumab and Bevacizumab in patients with recurrent Glioblastoma via Radiomic analysis on clinical MRI scans", Annual Conference on CNS Clinical Trials, 2021.
- Ruchika Verma, Ramon Correa, Virginia Hill, Niha Beig, Abdelkader Mahammedi, Marwa Ismail, Anant Madabhushi, Pallavi Tiwari, “Radiomic features from enhancing tumor on pre-treatment multiparametric MRI scans are predictive of response to chemo-radiation therapy in Glioblastoma and are associated with histological phenotypes”, in The International Society for Magnetic Resonance in Medicine (ISMRM), 2019, Montreal, Quebec, Canada.
- Sukanya Iyer, Marwa Ismail, Benita Tamrazi, Ashley Margol, Ramon Correa, Prateek Prasanna, Niha Beig, Ruchika Verma, Volodymyr Statsevych, Anant Madabhushi, Pallavi Tiwari, "Gradient-entropy based radiomic features to predict molecular sub-types of pediatric Medulloblastoma on Gadolinium-enhanced T1w MRI", in The International Society for Magnetic Resonance in Medicine (ISMRM), 2019, Montreal, Quebec, Canada.
- Niha Beig, Prateek Prasanna, Ruchika Verma, Virginia Hill, Vinay Varadan, Anant Madabhushi, and Pallavi Tiwari, "Radiomic features of Glioblastoma on pre-treatment Gd-T1w MRI are predictive of response to chemo-radiation therapy and associated with AKT and apoptosis pathways", in Proceedings of the Journal of Neuro-Oncology, 2018, New Orleans, Louisiana, USA.
